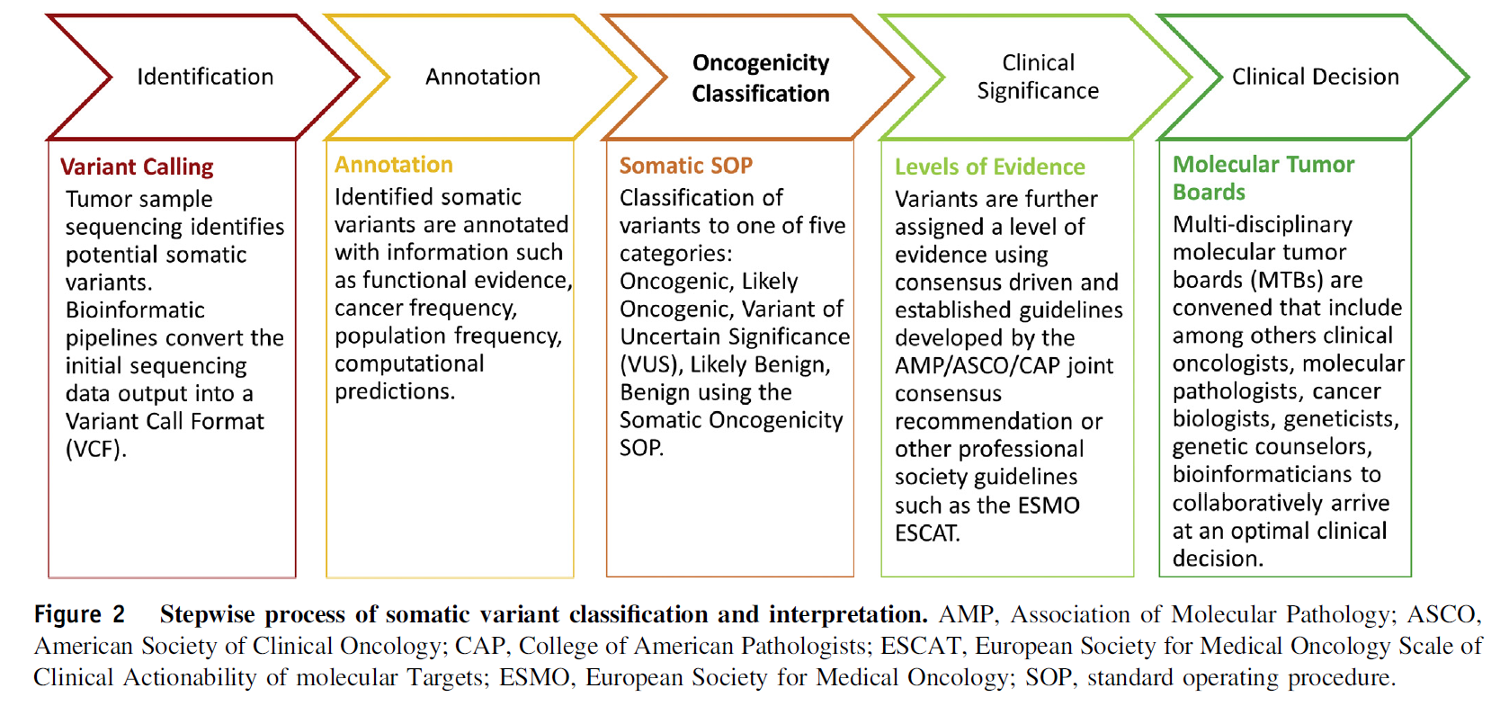
Classification of Variants Based on Oncogenicity Calculator js-html written by Dr. Thomas Schneider, Emory Pathology
Gnomad: Population database (not always germline)
The gnomad database is not user-friendly. You will need to figure out the specific format (see the Examples on the webpage) to perform a search. Or you can go to Varsome first and get the variant fromat from there.Cancer Hotspot: Somatic mutations associated with cancer
VarSome: the human genomic variant search engine
COSMIC: Catalogue of Somatic Mutation In Cancer
My Cancer Genome®: Genetically Informed Cancer Medicine
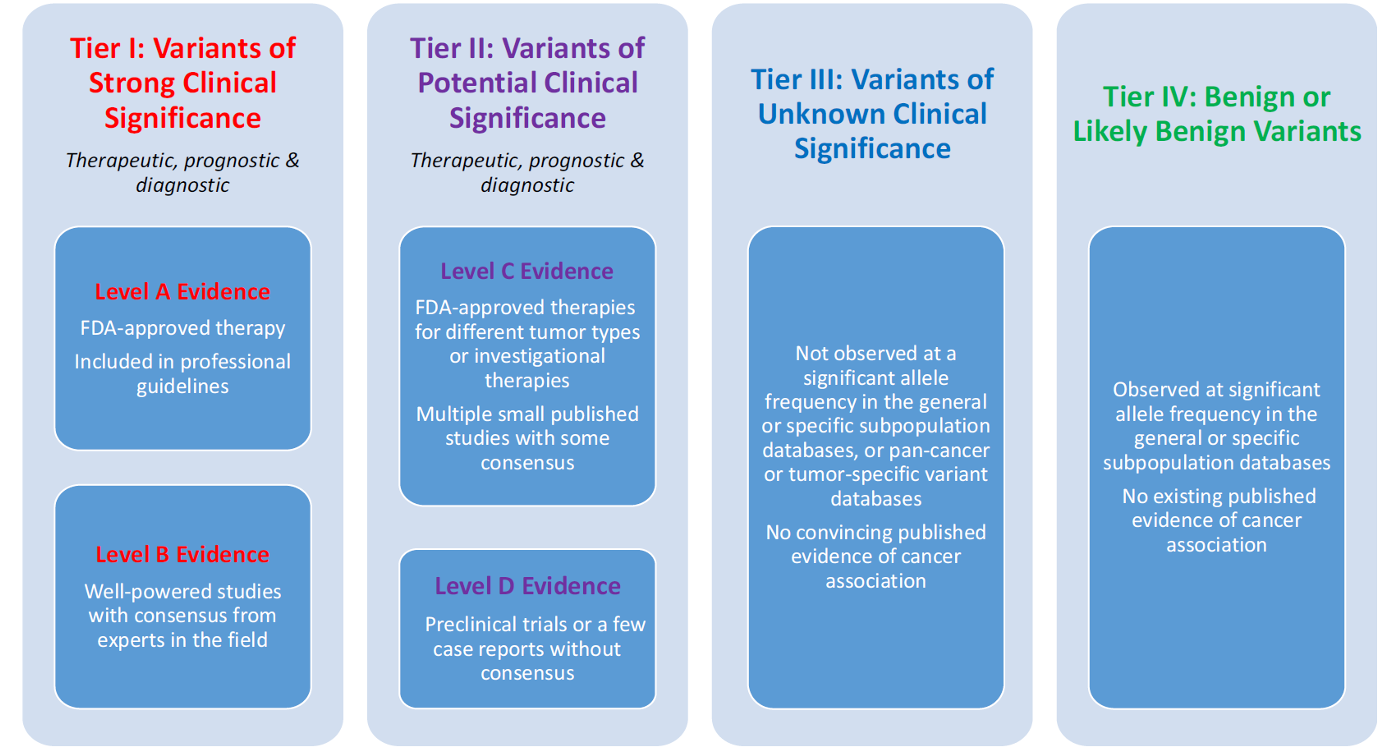
Copyrighted Figure, displaying here for educational purpose only.
See reference below.
Reference: Li, M. et al. Standards and Guidelines for the Interpretation and Reporting of
Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular
Pathology, American Society of Clinical Oncology, and College of American Pathologists.
J Mol Diagn. 2017;19(1):4-23.
PMID: 27993330
OncoKB: MSK's Precision Oncology Knowledge Base
CKBBoost: The Clinical Knowledgebase, Powered by The Jackson Laboratory
Back to the top Back to playground
This Website is Built by: Linsheng Zhang, MD, PhD. Emory University Pathology. September 2022.